Since the onset of the COVID-19 pandemic, the spike protein of the SARS-CoV-2 virus has been considered the primary component in its virulence and transmissibility. The dozens of mutations found on the spikes of the coronavirus' variants—particularly variants of concern (VOC)—are not only associated with their increased contagiousness but also with their immunoevasive abilities. Now, scientists have identified other proteins that are crucial in the severity of the disease.
A new multi-institutional study has found that certain "accessory" or nonspike genes, and their mutations, affect the pathogenesis of the SARS-CoV-2 virus. The animal study also learnt the spike protein influenced the severity of infection only in a few variants of the pathogen.
"Our work has revealed that the accessory proteins contribute to SARS-CoV-2 pathogenesis and the nonspike mutations in variants can contribute to replication of SARS-CoV-2 and pathogenesis in the host. This work suggests that while spike mutations may enhance receptor binding and entry into cells, mutations in accessory proteins may alter clinical disease presentation," wrote the authors. The findings were published in the journal PNAS.
Beyond Spike Protein
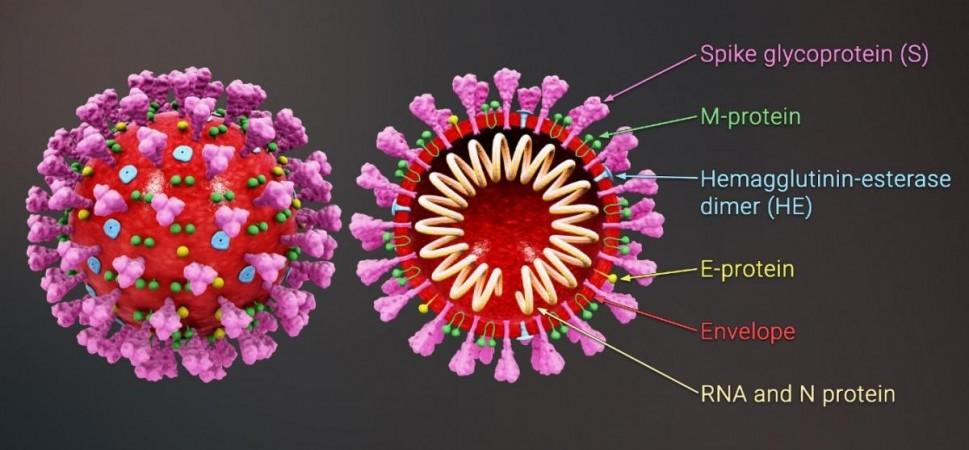
The SARS-CoV-2 virus possesses three types of genes: those associated with its replication, those that give it its structure, and accessory genes that are involved in other functions. While mutations in the spike protein—the hook-like structure that the SARS-CoV-2 uses to invade cells—is considered the key component in the virulence of COVID-19's variants, the impact of alterations in accessory genes has remained largely unknown.
Through the current study, the team aimed to ascertain what the functions of these accessory genes are. Therefore, they synthesized viruses that lacked each of four accessory proteins: ORF3a/b, ORF6, ORF7a/b, and ORF8. These viruses, along with the original virus, were used to infect different groups of mice. Following this, they analysed how each of these viruses affected the mice they infected.
Affecting Pathogenesis
It was found that the absence of the ORF3a/b gene in a virus resulted in milder infections when compared to the original SARS-CoV-2 strain. In contrast to mice infected with the original virus, the mice infected with this engineered variant lost lesser weight and had lower levels of the virus in their lungs.
This suggested that the ORF3a/b gene possibly contributes to restricting immune response to the infection or making additional copies through viral replication. Further experiments indicated that the gene may also play a role (within the virus) in activating the body's innate immune system, the first line of immune defense that responds to the germs entering the body.
However, the mice infected with the manufactured virus lacking the ORF8 gene were found to be sicker than those infected with the original SARS-CoV-2 strain. The inflammation in their lungs was significantly higher. According to the authors, ORF8 appears to influence the immune response within the lungs.
"By inhibiting the immune response, ORF8 helps the virus to replicate more in the lungs which worsens infection. When removed, it allowed the immune system to fight back harder," said Dr. Matthew Frieman, corresponding author of the study, in a statement.
Limited Commonality In Spike Function
The team's next goal was to determine the importance of the spike protein in the severity of the infection caused by different variants of the SARS-CoV-2 virus. For this, they replaced the spike gene of the original virus with that of its mutants (alpha, beta, gamma, or delta). Next, they infected mice and cells with these synthesised viruses and examined their replication and invasion of cells.
It was discovered that the severity of infection is determined by the spike protein only for a few of the variants. For example, the gamma variant's ability to infect and replicate was weaker when compared to other variants. According to the scientists, it is likely that mutations in genes outside the spike—specifically in the ORF8 gene—play a role in weakening this version when compared to others.
Dr. Mark T. Gladwin, Vice President for Medical Affairs at the University of Maryland (one of the universities contributing to the study), emphasised that future studies must focus on nonspike genes. "We need to learn more about the role of accessory protein mutations in COVID-19 infection, especially as new variants and subvariants keep emerging where these other proteins may play more of a starring role," he stressed.

















