With an estimated 9.6 million annual causalities, cancer is the second leading cause of death globally. While cancer treatments such as chemotherapy and immunotherapy help patients beat the disease and enter remission, certain immune-associated factors serve as hurdles in their ineffectiveness among some. Neoantigens, peptides produced by cancer cells, are one such example. Now, scientists have developed a new machine-learning algorithm that can identify which neoantigens are detected by the immune system.
In a multi-institutional study, researchers described a new machine learning technique, pMTnet. The algorithm was found to predict with high accuracy the neoantigens that were recognized by the immune cells known as T cells, and those that evaded them. New avenues in the prediction of cancer outcomes and potential effectiveness to immunotherapies may become explorable as a result of the technique.
"Determining which neoantigens bind to T cell receptors and which don't has seemed like an impossible feat. But with machine learning, we're making progress," said Dr. Tao Wang, senior author, in a statement. The findings of the research were published in the journal Nature Machine Intelligence.
Need for Better Tools
Different neoantigens are expressed on the surface of cancer cells depending on the mutations in the cell's genome. Immune T cells—that seek invasive pathogens and signs of cancer—are capable of identifying some of these neoantigens. Thus, they are neutralized by the immune system. However, some neoantigens can evade detection by T cells, which results in the growth of cancer.
Immunotherapy, which mobilizes an individual's immune system against cancer, can be greatly benefitted by a method that can predict which neoantigens are detected by T cells. It can aid researchers in developing better T cell-based therapies or in predicting the effectiveness of other forms of immunotherapies. The engineering of oncological measures such as personalized cancer vaccines may also gain by the availability of such information.
Tianshi Lu, first co-author of the study, explained, "For the immune system, the presence of neoantigens is one of the biggest differences between normal and tumor cells. If we can figure out which neoantigens stimulate an immune response, then we may be able to use this knowledge in a variety of different ways to fight cancer."
However, the existence of different neoantigens numbers in tens of thousands. Current methods of anticipating which of these numerous neoantigens can induce T cell responses are technically challenging, time-consuming, and most importantly, expensive.
Training the Algorithm
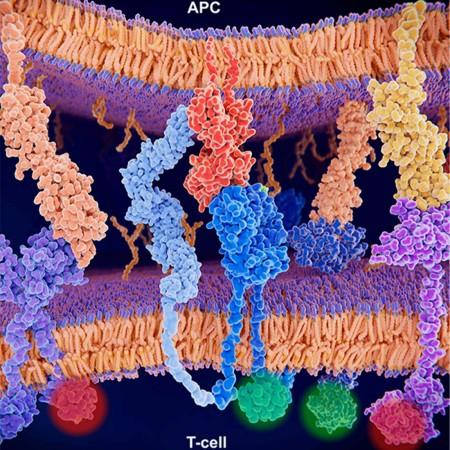
In order to find a technique that overcomes the challenges of predicting T cell response against neoantigens, the authors turned to machine learning. For this, they trained a deep learning-based algorithm that they named pMHC–TCR binding prediction network (pMTnet). Data associated with known binding or nonbinding combinations of three different components were used for training the algorithm.
The three constituents were: neoantigens; major histocompatibility complexes or MHCs (proteins that present neoantigens on cancer cell surfaces); and the T cell receptors (TCRs) that are responsible for the recognizing of neoantigen-MHC complexes. Following this, the algorithm was tested.
Accurate Prediction
pMTnet was evaluated against a dataset that was created from 30 different studies that had experimentally identified binding or nonbinding neoantigen T cell-receptor pairs. This analysis revealed that the newly-developed algorithm demonstrated a high level of accuracy.
Next, the team the used pMTnet to learn more about the neoantigens listed in The Cancer Genome Atlas, a public database that contains data from over 11,000 primary tumors. The new tool revealed that neoantigens usually initiate a powerful immune response when compared to tumor-associated antigens.
The algorithm also helped predict patient associated outcomes. This included which patients had better responses to therapies for immune checkpoint blockade and had better overall survival rates.
Highlighting the potential of the algorithm, Dr. Alexandre Reuben, corresponding author of the study, concluded, "As an immunologist, the most significant hurdle currently facing immunotherapy is the ability to determine which antigens are recognized by which T cells in order to leverage these pairings for therapeutic purposes. pMTnet outperforms its current alternatives and brings us significantly closer to this objective."














