The world has been battling COVID-19 for nearly 18 months now. As the pandemic has progressed, the SARS-CoV-2 coronavirus has also mobilized new troops—variants of concern (VOC). These evolved forms of the virus carry mutations that escape immune responses induced naturally or through vaccination. Now, a new study reveals the key mutation-led structural changes in the variants that enable them to evade the immune system.
According to scientists from Scripps Research Institute, the mutations in the variants are clustered in one particular area of the spike protein known as the receptor-binding site (RBS). The team conducted a comprehensive analysis of how important types of neutralizing antibodies (nAbs) bound with the original virus and its new forms. This helped them understand how the mutations helped the variants impedes the binding process
"This work provides a structural explanation for why antibodies elicited by COVID-19 vaccines or natural infection by the original pandemic strain are often ineffective against these variants of concern," said Dr. Ian Wilson, senior author of the study.
Variants With Potent Mutations
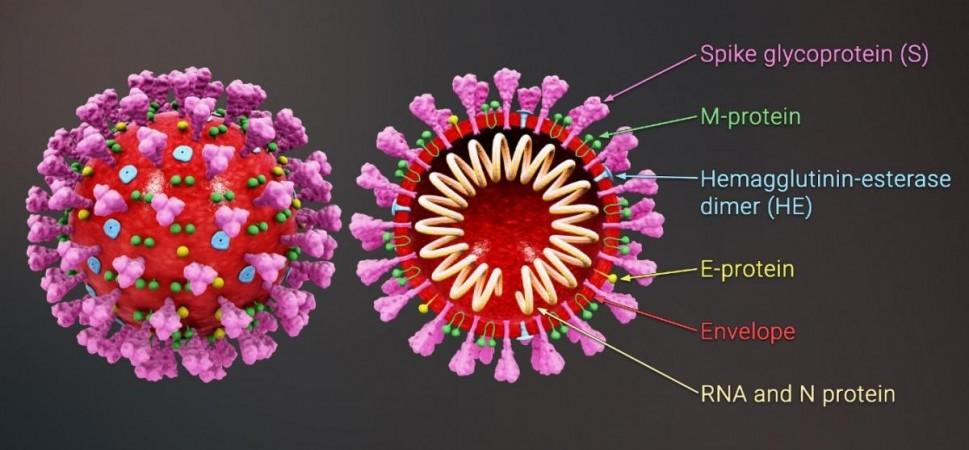
As of May 2021, four variants have been declared as VOCs by the World Health Organization (WHO)—P.1 (Brazil), B.1.617 (India), B.1.351 (South Africa), and B.1.1.7 (UK). Some of the VOCs, such as B.1.617 are far more contagious than the original Wuhan virus. Several recent studies have also demonstrated that protective antibody responses induced against the original strain (through natural immunity via infection or through vaccination) have decreased effectiveness in negating these variants.
Mutations on the SARS-CoV-2 spike protein (the hook-like structure that the virus uses to invade human cells) are said to be the source of their increased transmissibility and immuno-evasive abilities. The mutations are found in the RBS of the spike, which is the area where the binding with the host cells takes place.
Investigating Binding Ability
For the study, the authors primarily focused on three mutations in the spike: E484K, K417N, and N501Y. Either individually, or in combination, all three mutations are found in most of the major variants of the SARS-CoV-2 virus. Representative antibodies from major classes of nAbs that target the area in, and surrounding, the RBS, were evaluated. It was found that when the mutations were present, several of these nAbs lost their capacity to efficiently bind with the virus and neutralize it.
Next, the authors used structural imaging techniques to map the relevant section of the RBS at an atomic-scale resolution. This was done in order to understand the manner in which the mutations affect the areas where the antibodies would normally bind and neutralize the coronavirus.
It was gleaned that while the three crucial mutations greatly affect the antibodies' ability to bind within the RBS, they do not affect other vulnerable sites outside the RBS. The team also demonstrated that when virus-neutralizing antibodies targeted two areas outside the RBS, their ability remained largely unaltered by the trifecta of mutations.
Need to Exploit Vulnerabilities
The findings of the study illustrate that the antibody response against the RBS in the original Wuhan—that are potent to it—is severely diminished or nullified against some variants. Therefore, the need for updated COVID-19 vaccines to mitigate this hurdle may arise.
The study also suggests that future antibody-based treatments and vaccines may provide wider protection against the novel coronavirus and its variants, by targeting parts of the SARS-CoV-2 outside the RBS. This can be done either by employing or inducing antibodies against the site, the authors said. With the high likeliness of the virus becoming endemic in human populations, the team highlighted that broad protection against variants is essential.
"An implication of this study is that, in designing next-generation vaccines and antibody therapies, we should consider increasing the focus on other vulnerable sites on the virus that tend not to be affected by the mutations found in variants of concern," remarked Dr. Meng Yuan, co-lead author of the study.

















