While the world battles the COVID-19 pandemic on one side, several countries such as India are battling concurrent epidemics of opportunistic pathogens wreaking havoc among patients and survivors of the disease. These outbreaks have therefore brought attention to the potency that these otherwise harmless microorganisms possess. Now, scientists have reported that a common yet lethally sly bacteria—Pseudomonas aeruginosa—can be rendered nearly useless by inhibiting a mechanism that confers it with virulence.
According to a new study by researchers from the University of Geneva, a newly identified regulator (or an RNA helicase) of gene expression of P. aeruginosa—which has developed resistance to several antibacterials—is responsible for its pathogenicity. The study found that when the regulator was 'switched off', it drastically reduced the bacteria's ability to infect.
"Pseudomonas aeruginosa has an RNA helicase whose function was unknown, but which was found in other pathogens. We wanted to understand what its role was, in particular in relation to the pathogenesis of the bacteria and their environmental adaptation," said Dr. Martina Valentini, lead author of the study, in a statement.
An Opportunistic Pathogen
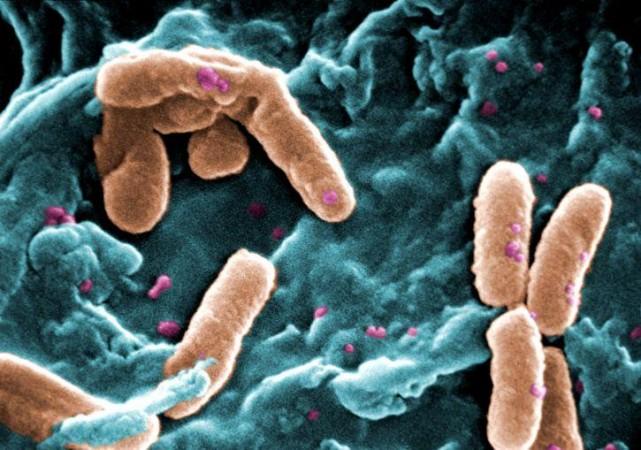
P. aeruginosa is a widely prevalent rod-shaped encapsulated bacterium that causes diseases in both plants and animals, including humans. It is found in varied ecological recesses; from stagnant water to roots of plants to hospitals. Being versatile and opportunistic, it can give rise to severe and chronic illness that often prove fatal. Individuals who are immunocompromised and are battling other acute infections serve as P. aeruginosa's primary targets.
The bacterium is also responsible for causing co-infections and super-infections in hospitalized patients by colonizing crucial medical equipment and leading to graver infections such as pneumonia and sepsis syndromes. During the course of the COVID-19 pandemic, several studies have reported P. aeruginosa-caused infection in patients struggling with the SARS-CoV-2 virus infection. Most importantly, the pathogen's progressive resistance to antibacterial drugs has made its treatment increasingly difficult.
RNA helicases are enzymes that perform regulatory functions by moderating structural changes of RNA molecules. These regulating proteins are present in the genomes of nearly all living organisms such as plants, animals, and microorganisms. Interestingly, they acquire properties based on the type of organism they are found in. DEAD-box RNA helicases—a family of helicases—that P. aeruginosa also has, is largely understudied. For the current research, the authors zeroed in on two homologs of the identified helicase from the family—RhlE1 and RhlE2. While RhlE1 helps the pathogen survive in cold climates, RhlE2 affects functions that are directly involved in virulence and adaptation.
Rendering the P. aeruginosa Harmless
In order to understand the role of the regulating protein in P. aeruginosa's virulence, the researchers integrated molecular genetic and biochemical approaches. Dr. Stéphane Hausmann, first author of the study, explained: "In the absence of this RNA helicase, P. aeruginosa multiplies normally in vitro, both in a liquid medium and on a semi-solid medium at 37°C. To determine whether the infection capacity of the bacteria was affected, we had to observe it in vivo in a living organism."
Next, the authors continued the research using Galleria mellonella larvae, an insect model often used to study host-pathogen interactions. The innate immune system of the insect shares several similarities with that of higher animals such as mammals. Importantly, these larvae can survive between a varied temperature range of 5°C and 45°C; making them ideal for studying bacterial growth across different temperatures, including the human body's.
Three groups of larvae were examined for this phase of the study. The first group, which was infected with a saline solution, witnessed 100 percent survival of its population. In the second group that had been infected with a normal strain of P. aeruginosa, less than 20 percent of the larvae survived 20 hours post the bacterium's introduction. However, when the third group was exposed to P. aeruginosa that was stripped of the crucial RNA helicase gene, over 90 percent of the group survived. "The modified bacteria became almost harmless, while remaining very much alive," highlighted Dr. Hausmann.
Blocking Without Killing the Bacterium
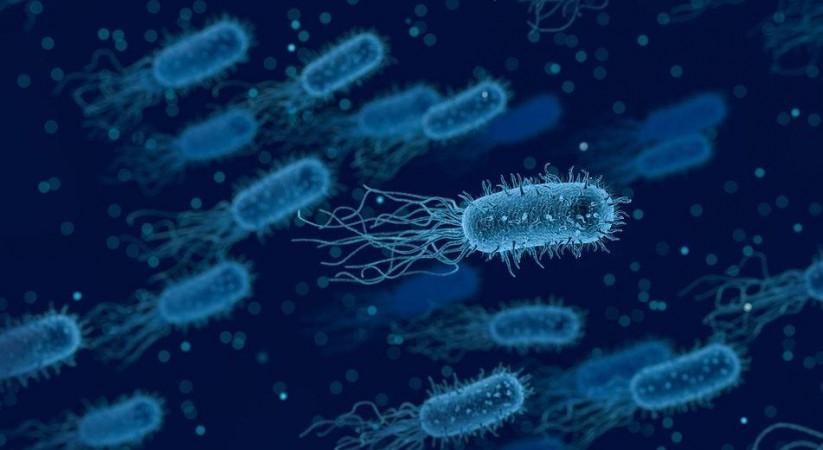
The findings of the study provide sufficient evidence to prove that the regulator protein indeed influences the production of several pathogenic factors in P. aeruginosa. "In fact, this protein controls the degradation of numerous messenger RNAs coding for virulence factors," stated Dr. Valentini. She emphasized that from the perspective of devising drug strategies, 'switching off' the bacterium's virulence factors instead of attempting to destroy it entirely can enable the immune system of the host to naturally neutralize the virus. It will also significantly reduce the chances of the pathogen developing resistance to drugs.
"Indeed, if we try to kill the bacteria at all costs, the bacteria will adapt to survive, which favours the appearance of resistant strains," stressed Dr. Valentini. The team has already begun screening existing drug molecules in order to ascertain whether any of them possess the ability to selectively negate the regulatory protein. They are also investigating the mechanism of inhibition based on which an effective therapeutic strategy can be developed.

















